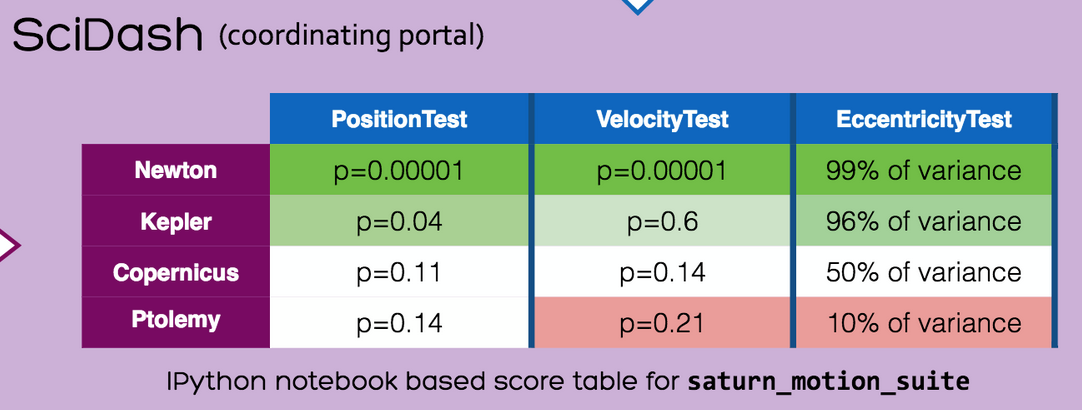
How to Test Stuff If You Do Computational Neuroscience
Superficially, this post is about SciUnit, a Python library. You can use it to test your computational neuroscience models written in any Python simulator - NEST, NetPyNE, Brian, and what not. We went for NetPyNE today.
This post is also about testing brain models in general. In fact, this post is about testing your *scientific models* - be they neuroscience models, a model of the solar system, or a model of the spread of the virus in a population. Consider the testing of the model of human brain circuitry presented here a case study for the broader topic of *automated* testing of scientific theories.
Introduction
I spent the last few months looking into the code behind (neuro-)scientific papers.
The code tends to look (at least when it comes to computational neuroscience) like you'd expect it to look - a one-off script that was never meant to be shared, reused, or read really, by anyone not on the team.
The closest analogy, if you come from software development, would be the scraping script, - the code we'd like to lie around in case we need to rerun it at some point, but also the code that we don't subject to too much scrutiny. It's not the code we'll "live in" in the next few years, it's not the code that someone will have to inherit, it's the code used to produce the results, the results we'll proceed to copypaste into the pdf.
In this post I'll share my experience with working on transforming a particular paper presenting a scientific model into something other than a one-off script. I'll describe why we might want to do this, and I'll describe how we might want to do this.
This is the "best practices" post, or, as I much prefer it, "a post aimed to help you acquire a better sense of what might be a good idea in certain contexts".
Most "good ideas" in the context of scientific code match the ideas we consider good in software development, some, however, do not, and this is what we'll focus on.
Reproducibility (not to be confused with Replicability!)
Let's begin with a nice categorisation, borrowed from the "Re-run, Repeat, Reproduce, Reuse, Replicate: Transforming Code into Scientific Contributions" paper, of the properties the code accompanying your paper might have.
- Rerunnability (R1) - you can rerun your code, whatever the output.
For example, if you used Python 2 - it needs to be explicitly declared, so that the person rerunning your program knows what environment to use. Freeze your dependencies. - Repeatability (R2) - the output of your code should be the same every time.
For example, if there is randomization in your code - set the seed of therandommodule. Not doing this introduces unnecessary point of uncertainty to your results - does my output slightly differ from the output stated in the paper because I uncommented the wrong parameter, or because of the randomized values? - Reproducibility (R3) - another researcher should be able to exactly reproduce your results in an automated, standardized, and self-verifiable manner.
Reproducibility implies both rerunnability and repeatability, but it has other researchers in mind. Your code, data used in your code, and results of the run of your code should be in the open access. Your code should be self-verifiable (tests). An outside researcher should be able to tell which branch/commit you used in your paper. Most importantly - results should be presented in the automated, standardized format. See "Ten Simple Rules for Reproducible Computational Research" for more. - Reusability (R4) - your code should be comprehensible, comment it and document it.
A person (say you in 2 years) should not only be able to rerun your code, but they should also be able to understand and reuse your code in a project of their own. - Replicability (R5) - a person reading your paper should be able to replicate your code based on the information provided in the article alone, and they should be able to get similar results.
"Replicability" here is used in the usual scientific sense.
Rerunnability (R1), Repeatability (R2) and Reusability (R4) are standard concerns any software developer is probably aware of.
Replicability (R5) is a standard concern any researcher is probably aware of.
Reproducibility (R3), however, is neither, and that's what we'd like to discuss.
As is frequently the case with subrigorous definitions, the strict division is not important per se. The main takeaway is that, when it comes to the scientific code, there may exist concerns that are captured neither by the software development best practices, nor by the best practices of research.
Reproducibility as described here pertains exclusively to the scientific code.
Reproducible code automatically (without requiring additional efforts from the reader) reproduces the main results of the paper (tables and graphs) in the standardized manner.
There's a famed article in The Atlantic suggesting Jupyter Notebooks might be the future of the scientific paper.
Jupyter may be far from becoming the default media in research (if that's even a worthy pursuit), however Jupyter Notebooks already give us a good hunch for what the reproducible code should look like: it should be possible to put your code into the executable notebook environment, have someone run it without changing any parameters, and have them see the output similar to that published in your paper.
When the paper is not Reproducible
Let's use a particular paper for an example of what non-reproducible code looks like.
The paper we'll use describes the process of converting the model of the 1mm2 sensory cortical network from NEST to NetPyNE (both are biological neural network simulators), and of rescaling the resulting NetPyNE neural network while preserving its first- and second-order statistics.
I advise you to throw an eye over these links - in particular github repositories, and to read through the Jupyter notebook.
| The paper "NetPyNE Implementation and Scaling of the PDCM Model" | link (mit.edu) |
| The original code (non-reproducible) behind the paper | link (github) |
| The transformed code (reproducible) | link (github) |
| Jupyter notebook with the transformed code | link (jupyter) |
Take a look at the original code. In order to reproduce the results in the paper (given in paper's tables and in paper's text), one would have to manually change these (and other) parameters in the original code:

For example, in order to reproduce one of the tables in the paper (Table A.1, given below), we have to change the parameters 10 times (for each scaling of the network), and manually calculate the relative difference score (given in parentheses).
[omg picture disappeared!]
The NEST trial row shows the stats of the NEST model (Gewaltig & Diesmann, 2007) the paper tried to recreate.
The 100% row shows the stats of the full-scale NetPyNE model.
The 80%, ..., 1% rows show the stats of various scalings of the NetPyNE model.
[The "NetPyNE Implementation and Scaling of the PDCM Model" paper]
In order to reproduce any other table given in the paper, we'd have to repeat the procedure.
Notice that the original code is good - it does reliably produce the results mentioned in the paper. However it lacks automatization (we can't reproduce the entire paper in one run), standardization (the researcher with an idea for a different rescale function cannot simply implement it, and rerun the code to see how it compares to the original rescale function), and self-verification (there are no tests, and you wouldn't know whether the results of the run are alright without comparing your output to the output in the paper, line by line).
The transformed code uses the SciUnit framework - SciUnit validates your scientific model against data - in a way that makes your code reproducible, and, thus, ready for "Jupyterification". If, in order to reproduce the results in the paper, the original code requires the researcher to manually adjust the parameters, record the results, and calculate the score on their own, transformed code has this entirely automated.
For example, the table_a1() function reproduces Table A.1.

SciUnit, so far, is the only package that aims to rectify the aforementioned shortfalls (with the gotcha for self-verification, as I'll later explain).
Writing your code like you write your paper: follow the tables
SciUnit encapsulates your code in a structure similar to that of a scientific paper.
You are required to create Models, and Tests that probe the features of those models.
An interesting question is what constitutes a scientific model.
Suppose we're considering a model of the human cortex, and we're interested in synchrony. When we adjust the weight of nicotinic synaptic connections - is this the new model of the human cortex, or is this the same model, tested with nicotinic agonists?
Translating this to the real-world terms, would you consider a person smoking a cigarette a person with the new model of the brain, or would you consider this person's brain a constant that can be tested with or without a cigarette affecting it?
In other words, do we run 1 test [get_synchrony()] on 2 models [brain_with_cigar, brain_without_cigar],
| 1 test, 2 models | |
| synchrony | |
| brain affected by cigar | 111 |
| brain not affected by cigar | 222 |
or do we run 2 tests [get_synchrony_with_cigar(), get_synchrony_without_cigar()] on 1 model [brain]?
| 2 tests, 1 model | ||
| synchrony before cigar | synchrony after cigar | |
| brain | 111 | 222 |
Notice how this question doesn't arise when we do a vanilla simulation (like we do in the original code behind the paper), we would simply adjust the weight of the connection to nicotinic receptors in our model, and run model.get_synchrony(). Did we just create a new model, or a new test case for our model? It can be seen as both, and we are not forced to make the choice.
The moment we have to choose is when we build a table to present our results in the paper.
And SciUnit, moulding your code into the structure of the paper, correspondingly has to force you into this choice.
I find it a good idea to avoid thinking about your tests and models in the real-world terms (did the model of my brain change after I smoked the cigarette?).
The result of your decision is the form of the table - think of what should be the rows of your table (these will be the models), think of what should be the columns in your table (these will be the tests), and what values you want in the cells of the table.
Conceptually this is similar to how one builds programming interfaces - you should never have to think whether this class is conceptually, or in the real world, similar to the other class - you should come strictly from the 'what behaviour do these classes share' standpoint.
SciUnit VS other testing frameworks, or
Testing your model ≠ testing your code
"When you construct a model you leave out all the details which you, with the knowledge at your disposal, consider inessential. Models should not be true, but it is important that they are applicable, and whether they are applicable for any given purpose must of course be investigated."
Georg Rasch (1960), Probabilistic Models for Some Intelligence and Attainment Tests
Your model is expected to fail some "tests".
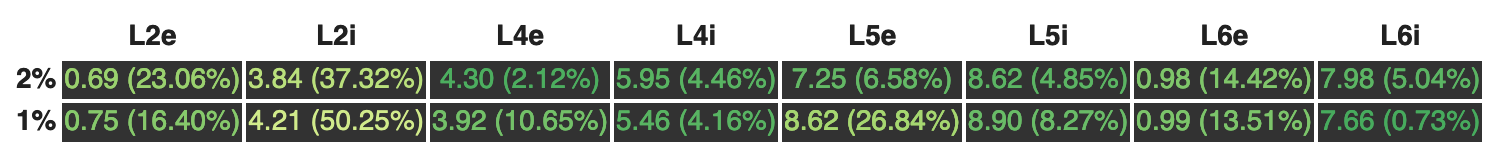
For example, returning to the PDCM paper, you see the 1%-scaled network resulting in the 46% relative difference score for the L2/3i cortical layer.
Model validation is defined as the comparison of predictions generated by the model to the experimental results (or to the perfect results, the 100%-scaled network in our case) in the context of the model's intended domain of applicability.
The 1%-scaled network runs for a couple of seconds on a single-core machine, the 100%-scaled network runs for hours. Considering our domain of applicability (neural networks that are not prohibitively expensive computationally), we note our 46% deviation from the ideal result, and we still consider the 1%-scaled model to be of value.
[omg picture disappeared!]
[The "NetPyNE Implementation and Scaling of the PDCM Model" paper]
Previously I mentioned that SciUnit doesn't make our code self-verifiable.
Let's take a look at the output of SciUnit that aims to display the last 2 rows of the table above:
[Jupyter notebook using the transformed code]
1%-scaled model's mean firing rate in the population L2i (cortical layer 2, inhibitory) is off from the ideal result by 50.25% (which is close to the paper's 46% score), and all we get from this diversion from the perfect result is a yellowish text color.
As we discussed, we are happy with this result, and our model validation must not fail because of this, and SciUnit doesn't raise any errors.
Even though our 1%-model doesn't match the ideal results - it does match our (model developer) expectations!
A standard testing framework tests your scientific model against your expectations.
SciUnit (or any other model validation framework, or tables in your paper) tests your scientific model against experimental data (or against perfect results).
Again - SciUnit tests your model against experimental results. Your code may be perfectly valid, and still produce the 50.25% (or 100%, or 200%) error - you just coded a scientific model that is not applicable to this domain!
SciUnit doesn't have an opinion on whether the 50.25% score is acceptable for your model in this domain - it's up for you (the developer) to decide whether this score is acceptable, and to hardcode this into your model, and that's where traditional testing frameworks come in.
SciUnit should be accompanied by standard software tests, implemented, say, with unittest, and they should look somewhat like this (this is pseudocode):self.assertEqual(table.get('1%', 'L2i'), 50.25).
This is what self-verification is - not the colored cells in your table, not the scientist throwing an eye over the table and deciding whether the output looks reasonable - but the boolean, hardcoded developer expectations.
In this way, SciUnit is not a testing framework in a traditional sense, and there is no good analogy for this in software development - in software development, our "ideal results" always match "developer expectations".
Beyond your own paper
The consequences of structuing your code like you structure your paper spread vast beyond easy jupyterification and easy reproduction of the paper contents.
You want to change the stastistical measure used from relative difference to the z-score? It's a one-liner.
You want to see how exchanging the neuron model used affects your results? Substitute the .mod file, and rerun the test suites. This will take hours, but hours of computational time, not of your time.
You saw the PDCM paper, but you want to implement the rescaling of a completely different neural network, say the spinal chord? Implement the spinal chord neural network, import the test suites someone else already created for you, and run them.
You have an idea for the better rescaleNetwork() function? Change your function, and rerun the test suites.
You have an idea for the better rescaleNetwork() function, and you want it evident that your rescaleNetwork() function is better than the one used in the original paper? Import the original paper's model, create the test suite that compares both models, and submit it to the SciDash portal, so that someone with an even better rescaling function (according to them!) can run against your test suite in the future.
Largely due to the lack of the standardization across the field, summary questions are the hardest to answer in scientific modelling, and SciUnit provides this standardization for us.